What is Giotto?¶
Giotto is a comprehensive and open-source toolbox for spatial data analysis and visualization. The analysis module provides end-to-end analysis by implementing a wide range of algorithms for characterizing tissue composition, spatial expression patterns, and cellular interactions. Furthermore, single-cell RNAseq data can be integrated for spatial cell-type enrichment analysis. The visualization module allows users to interactively visualize analysis outputs and imaging features. Giotto is applicable to a wide range of spatial technologies and platforms.
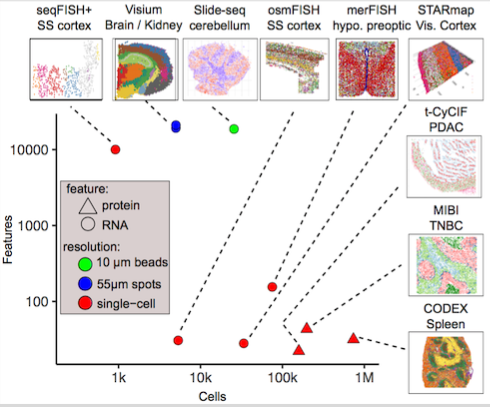
Description¶
Giotto provides a flexible framework for common single-cell processing steps such as:
Quality control
Normalization
Dimension reduction
Clustering and cell type annotation
To facilitate the analysis of recently emerging high-throughput, but lower-resolution spatial transcriptomic technologies, such as 10X Genomics Visium and Slide-seq, Giotto has 3 implemented algorithms for estimating the spatial enrichment of different cell types by integration of known gene signatures or single-cell RNAseq expression and annotation data.
Spatial information is retained through the formation of a spatial grid and/or a spatial proximity network, which is used to:
Identify spatial genes
Extract continuous spatial-expression patterns
Identify discrete spatial domains using HMRF
Explore cell-type/cell-type spatial interaction enrichment or depletion
Calculate spatially increased ligand-receptor expression in cells of interacting cell type pairs
Find interaction changed genes (ICG): genes that change expression in one cell type due to interaction with a neighboring cell type
Giotto provides a number of options to visualize both 2D and 3D data and the outcome of Giotto can be interactively explored using Giotto Viewer which allows you to overlay the obtained results with raw or additional images of the profiled tissue section(s).
Cite Giotto¶
Dries, R., Zhu, Q., Dong, R., Eng, C. H. L., Li, H., Liu, K., … & Yuan, G. C. (2021). Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome biology, 22(1), 1-31. <https://pubmed.ncbi.nlm.nih.gov/33685491/>
License¶

Authors and Developers¶
Author, Maintainer |
|
Qian Zhu |
Author |
Huipeng Li |
Author |
Rui Dong |
Author |
Guo-Cheng Yuan |
Author |